The population of microbes in our gut are a personal trait, like height or weight. The precise make-up of that population, its balance of different types of microbes, is significant as it can have major beneficial or harmful effects on our health. Like many other traits, the composition of that population is partly due to nature—our genes create an environment welcoming to certain types of microbes—and nurture, the food we eat.
Given the importance of the composition of the microbial community to our health, it’s not surprising that there is a lot of interest in trying to figure out just how significant are the contributions of nature and nurture. Comparing genetically identical mice with a drastically simplified community of microbes in their gut, researchers found that changes in diet led to predictable changes in community structure. However, comparing genetically distinct strains of laboratory mice, a different group of researchers found that the subtle genetic differences between two inbred lines of mice led to measurable differences in the gut microbial community (see part I).
The roles of nature and nurture can be probed further by comparing the microbial community of different, but related, types of animals. What if two animals are closely related, but have different diets? What if they are only distant relatives, but have similar diets? An extreme example of the latter case is a comparison of the intestinal microbes of the tammar wallaby and the cow (see part II). While the comparison is informative, these two mammals are about as distantly related as possible, so a closer comparison is useful.
In a recent study from Washington University at Saint Louis, a group led by Jeffrey Gordon attempted to compare a family tree of mammals, a family tree of the microbes that lived in their guts, the diets of those mammals, and the genetic abilities of their microbes. Any four-way comparison gets complicated pretty quickly, but there are some clear facts that emerge from the fog of statistics. These facts emphasize some basic biology of microbes, as well as telling us something about how mammals and their microbes co-evolve.
A family tree of the mammals comes out pretty much the same, regardless of whether you use genetic data or anatomical differences:
Looking at this family tree, there is little convergence between relatedness and diet. For instance, elephants, capybaras, gorillas, horses, and rabbits are all “hindgut-fermenting herbivores,” with a specific method of digesting cellulose—and yet, are found on different branches of the tree. Sheep, hyraxes, kangaroos, colobus monkeys, and pigs are all “foregut-fermenting herbivores,” with an anatomically distinct way of digesting plant matter—but again, they are only distant relatives. One can find groups, such as the primates, comprising foregut-fermenters, hindgut-fermenters, carnivores, and omnivores. So, genetics on a deep level does not determine diet.
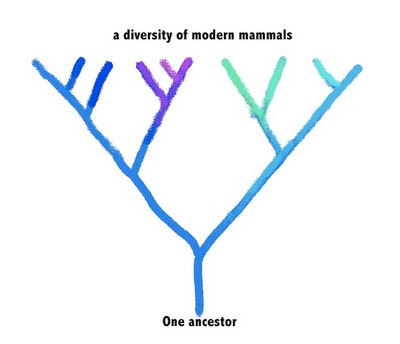
If genetic heritage doesn’t determine diet, does it at least determine something about a mammal’s intestinal microbial community? To approach this question, you can imagine a goodly branch of that mammalian family tree. A diverse bunch of modern mammals evolved from a common ancestor:
We obviously don’t know anything of the DNA sequence of their common ancestor, but we can see the genetic similarities of the modern animals, and work backwards to predict what their ancestor was like, as well as which modern animals are most closely related. If the ancestor had a certain genetic composition—symbolized by the color blue—then its descendants would have evolved variations on that genetic composition—symboized by variations in the hue in the modern beasts.
Now, if genetic heritage determines an animal’s gut microbes, then the genetic character of that community should show a pattern of evolution sort of like the mammalian family tree:
This is a trickier tree than the previous one. Rather than looking at a lineage of organisms, we are looking at the composition of a community of thousands of species. We can make an analogy with the white pages of a phone book: in this tree, we are looking at how the content of a community’s phone book changes over time. (Also, it’s important to remind ourselves that we don’t know what the original community was like—we only have the present one, and we try to work backwards.)
So, what did Gordon’s group find? If mammals and their intestinal microbes co-evolved, then we could examine a diversity of modern mammals and the microbial communities in their poop—which is just what they did. We’d expect related mammals to have related communities. Lining up the tips of the family trees (that is, just currently living animals and the microbes in their poop), we’d see something like this:
There was good reason to expect this result, as similar patterns of co-evolution are seen with insects and their symbiotic bacteria. Co-evolution is common. But here’s what they found:
What to make of this? Obviously, genetic heritage has very little to do with the community structure of an animal’s intestinal microbes. However, Gordon’s group looked at the same data again. This time, instead of looking at mammals by their ancestry, they considered their diet:
Aaaaah, much better! So, it seems like diet—nurture, in our nature/nurture dichotomy—is a much better predictor of a mammal’s microbial community. (This is a much simplified version of the data presented; my apologies to Gordon et al, who studied 33 mammal species and hundreds of microbial species. It’s also useful to note that in several instances they looked at the communities of different individuals of the same species, and found little variation between them.)
Interestingly, despite the differences in community structure between mammals, all mammals have pretty much the same “core” community of microbes. To return to the metaphor of a phone book, any big city in this country will have roughly the same collection of names, it’s just that one city will have a lot more Schmidts and Webers and Mullers and another will have many more Vasquezes and Diazes and Gonzaleses.
Although there was a decent correlation between mammalian diet and community composition in Gordon’s data, his group found an even more significant relationship. The best correlation was not between mammalian diet and community composition. It was between mammalian diet and what genes were present in the microbial community.
Let’s consider that phone book analogy again. If I wanted to find somebody to turn some wheat into flour, I probably wouldn’t turn to the white pages and call up somebody named Muller or Miller. (Heck, I wouldn’t say I’m a good horticulturalist because I’m named Appleman.) Trades are not inherited with family names, and this is true in the microbial world as well. So, if you’re a vegetarian mammal looking for a microbe to turn your b-glucosides into monosaccharides, you can’t assume that you’re looking for Brevibacter—all you care about is whether the organism has the genes for doing the job.
Microbial genomes—the collection of genes a given species has—are incredibly fluid. Genes can disappear from one genome in the space of a few generations, and can establish themselves in a new genome in an equally short time. Genes can travel with relative ease between bacterial species that are barely related to each other. (There are many mechanisms for doing this, and it makes firmly describing a microbial species next to impossible.) So, when a hyrax feeds its intestinal microbes a blend of grass and insects, it is (unwittingly) selecting a specific assemblage of genes, not a specific community of microbes.
These results raise some interesting questions about evolution. The community of microbes that lives in a mammal’s gut—its microbiome—can easily be viewed as an organ like the lungs or kidneys. However, its inheritance and evolution are quite distinct from that of the rest of the animal. As Gordon’s group suggests, it would be fun to compare a lot of different carnivores, and try to extrapolate just when differences in their microbiomes evolved. Do such evolutionary jumps correlate with events such as continental drift or other significant milestones? Are the changes gradual or rapid? Can one community be dislocated by another? Just how fluid are the communities in the gut?
These questions bring us back to the tammar wallaby and its oddly non-methanogenic digestion. Interestingly, in this study, the microbiome of a kangaroo clearly grouped with other foregut-fermenting mammals, such as sheep and gazelles. It was an outlier in that group, but still clearly part of it and not some other group. Well, the tammar wallaby was found to have a small number of methanogens and acetogens in its gut, giving it at least some similarity to the sheep. The big difference in digestion (and flatus) may be traceable to the very small community difference of one organism with one interesting metabolic pathway, a factor that simply can’t be revealed by this sort of analysis. The situation is reminiscent of human societies, which can be hugely affected by a single individual—and knowing the name of that individual won’t tell you anything about how they affect society.
Brian D. Muegge, Justin Kucyznski, Dan Knigts, Jose C. Clemente, Antonio Gonzalez, Luigi Fontana, Bernard Henrissat, Rob Knight, and Jeffrey Gordon (2011). Diet Drives Convergence in Gut Microbiome Functions Across Mammalian Phylogeny and Within Humans. Science 332, 970-974.






No comments:
Post a Comment